Glossary
Cas
CRISPR associated protein. There are many different Cas proteins, corresponding to different types and subtypes of the CRISPR system present in different species. Class 1 CRISPR systems employ a complex of different Cas proteins to degrade foreign nucleic acids whereas Class 2 CRISPR systems employ a single large Cas protein.
Cas9
The Class 2 type II Cas protein first used to demonstrate how the CRISPR system could be used to mediate gene editing by engineering synthetic guide proteins targeted to specific sites. Cas9 remains the best characterised and widely used Cas protein. Cas9 causes blunt ended double stranded breaks at the target site (3 bp upstream of the PAM). The most commonly used variant of Cas9 is that derived from Streptococcus pyogenes, which has a PAM of 5’-NGG-3’ (where N is any base). Cas9 proteins derived from different species will have different properties and a different associated PAM. The CRISPR/Cas9 complex requires both crRNA and tracrRNA to form an active complex.
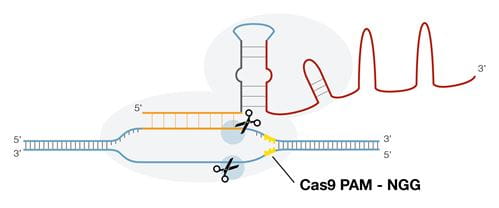
Ribonuclease complex of CRISPR/Cas9 system showing sgRNA bound to Cas9 protein and PAM sequence downstream from cleavage site.
Cpf1
A Class 2 type V Cas protein originally derived from Prevotella and Francisella bacteria, recently renamed by some as Cas12. Cpf1 is smaller than Cas9 and generate a 4 or 5 sticky ended overhang double stranded DNA break (as opposed to blunt ended as with Cas9). Cpf1 has a PAM of 5’-YTN-3’ (where Y is a pyrimidine, i.e. T or C, and N is any base) and cleaves further away from the PAM site. Cpf1 does not require tracrRNA to form a mature complex with crRNA. The Broad has patents over the CRISPR/Cpf1 system and has licensed these to its associated spin out Editas Medicine. On 19 April 2017 the EPO indicated its intention to grant the Broad's European Patent EP3009511 in respect of the CRISPR/Cpf1 system.
crRNA
CRISPR RNA. The sequence of RNA which guides the CRISPR/Cas complex to the target site for cleavage. Typically a crRNA sequence must be a minimum of 20 bp long for efficient site cleavage. In type II (Cas9) systems crRNA must interact with tracrRNA for to form a complex with the Cas9 protein. Other CRISPR systems such as Cpf1 do not require tracrRNA for crRNA to form a mature complex with the Cas protein.
CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats. Refers to the system of adaptive immunity discovered in bacteria and archaea which is characterised by the presence of these repeat regions interspaced with “spacer” regions of captured foreign DNA. The exact CRISPR mechanism differs from prokaryotic species to species – at present CRISPR/Cas systems are divided into two classes and six types – Class 1 is divided into types I, III and IV and Class 2 is divided into types II, V and VI.
C2c2
A Class 2 type VI Cas protein which targets single stranded RNA as opposed to DNA, recently renamed by some as Cas13. C2c2 is a RNA-guided, RNA-activated non-specific RNAse. Target sequence recognition is mediated by complementary non-G protospacer flanking sites (PFS). Upon binding of the C2c2 complex cleavage occurs through the C2c2’s non-specific RNAse activity at U residues in single stranded RNA regions of the target RNA, without requiring complementarity to the crRNA guide sequence.
Feng Zhang of the Broad published a widely publicised paper in Science which proposed a method (which he dubbed “SHERLOCK”, an acronym for specific high sensitivity enzymatic reporter unlocking) which uses the RNA binding and cleavage functionality of C2c2 as a sensitive and highly specific detection tool for viral RNA transcripts present in extremely low concentrations.
gRNA
PAM
Protospacer Adjacent Motif. The downstream presence of the particular PAM specific to the Cas protein employed in a CRISPR system is essential for binding of the Cas protein to the target DNA and cleavage of the target side – if the necessary PAM is not present the Cas protein will not bind. In bacterial systems, the PAM will be present on foreign DNA but not the bacterial CRISPR locus – by this means the CRISPR system is able to distinguish and selectively target foreign DNA for destruction.
sgRNA
Single Guide RNA. An RNA sequence which combines at least one crRNA and a tracrRNA in the one sequence, thereby simplifying the transfection and expression of the necessary components for the CRISPR system. This was one of the key contributions to the art in Doudna and Charpentier’s seminal Jinek 2012 paper.
tracrRNA
Trans-activating CRISPR RNA. A small hairpin loop RNA sequence which plays a role in the maturation of crRNA and formation of the crRNA-Cas9 complex (in type II CRISPR systems). In native systems tracrRNA is complementary to the repeat regions of the pre-crRNA transcript formed from the CRISPR region and forms a double stranded RNA duplex with the pre-crRNA, which is then cleaved to form a mature crRNA/tracrRNA hybrid which guides the Cas9 to the target locus. In sgRNA both the crRNA guide and tracrRNA minimal region necessary for Cas9 binding are present in the one sequence.

